3. Registration#
Correct for rigid/non-rigid movement
Perfom motion-correction (rigid or piecewise-rigid) to eliminate movement in your movie.
Run a grid-search across multiple sets of parameters.
Use quality metrics to evaluate registration quality.
%load_ext autoreload
%autoreload 2
from pathlib import Path
import os
import pandas as pd
import numpy as np
from mbo_utilities import imread
from matplotlib import pyplot as plt
import fastplotlib as fpl
import caiman as cm
import lbm_mc as mc
from lbm_mc.caiman_extensions.cnmf import cnmf_cache
from caiman.motion_correction import compute_metrics_motion_correction
import lbm_caiman_python as lcp
if os.name == "nt":
# disable the cache on windows, this will be automatic in a future version
cnmf_cache.clear()
3.1. Data path and batch setup#
We set 2 path variables:
data_path: input, path where you saved the output from the assembly step
This will set the global
parent_raw_data_path, so you can move the results anywhere you want as all results will be saved relative to this directory
batch_path: a results, can be anywhere you please, must end in.pickle
Note
This notebook assumes you saved scans as TIFF with join_contiguous=True so filenames are plane_N.tiff
The process for join_contiguous=False will differ and is not covered here.
In this demo we put our batch_path (which stores results) in raw directory, in its own folder.
This is helpful as you can move this folder anywhere you wish.
data_path = Path(r"C:\Users\RBO\lbm_data\processed")
batch_path = data_path / 'batch' / 'batch.pickle'
3.1.1. Create or load a batch set#
# Create or load a batch results file
# To overwrite (be very careful with this):
# batch_df = mc.create_batch(batch_path, remove_existing=True)
if not batch_path.exists():
print(f'creating batch: {batch_path}')
df = mc.create_batch(batch_path)
else:
df = mc.load_batch(batch_path)
# tell mesmerize where the raw data is
mc.set_parent_raw_data_path(batch_path.parent.parent)
WindowsPath('C:/Users/RBO/lbm_data/processed')
tiff_files = [x for x in Path(data_path).glob('*.tif*')]
tiff_files[0] # first file
WindowsPath('C:/Users/RBO/lbm_data/processed/plane_10_assembled.tiff')
3.1.2. Load a data file to examine#
file = tiff_files[1]
data = imread(file)
data.shape
3.1.3. View metadata#
lcp.get_metadata(filepath) works on raw scanimage files and files processed through lcp.save_as()
metadata = lcp.get_metadata(file)
metadata
{'num_planes': 30,
'num_frames': 1730,
'fov': [150, 150],
'num_rois': 4,
'frame_rate': 9.60806,
'pixel_resolution': [1.04, 1.0],
'ndim': 3,
'dtype': 'uint16',
'size': 18648189000,
'raw_height': 2478,
'raw_width': 145,
'tiff_pages': 51900,
'roi_width_px': 144,
'roi_height_px': 600,
'sample_format': 'int16',
'objective_resolution': 157.5}
dxy = metadata['pixel_resolution']
print(f"Pixel resolution: {dxy[0]} um/px, {dxy[1]} um/px")
Pixel resolution: 1.04 um/px, 1.0 um/px
# note this is only for a single ROI
fov = metadata['fov']
print(f"Field of View: {fov[0]}px, {fov[1]}px")
Field of View: 150px, 150px
fr = metadata['frame_rate']
print(f"Frame rate: {fr} Hz")
Frame rate: 9.60806 Hz
3.2. Registration parameters#
Parameter |
Description |
Value/Default |
|---|---|---|
|
Spatial resolution (pixel size in micrometers). |
|
|
Frame rate of the video (frames per second). |
|
|
Maximum allowed rigid shift in pixels for motion correction. |
|
|
Size of patches for motion correction. |
|
|
Overlap between patches for motion correction. |
|
|
Maximum allowed deviation for patches relative to rigid shifts. |
|
|
How to handle border values during motion correction. |
|
|
Flag indicating whether to perform piecewise rigid motion correction. |
|
|
Size of the Gaussian filter for smoothing the motion correction process. |
|
|
Flag to use bicubic interpolation for motion correction. |
|
The parameters are passed directly to caiman, this means you need to use the same exact names for the parameters and you can use all the parameters that you can use with caiman - because it’s just passing them to caiman.
The parameters dict for a mesmerize batch item must have the following structure. Put all the parameters in a dict under a key called main. The main dict is then fed directly to caiman.
{"main": {... params directly passed to caiman}}
3.2.1. Using metadata to assign parameter values#
Important
The goal here is to get an approximate neuron size in microns.
This value is used as the basis of the patch and max_shifts parameters.
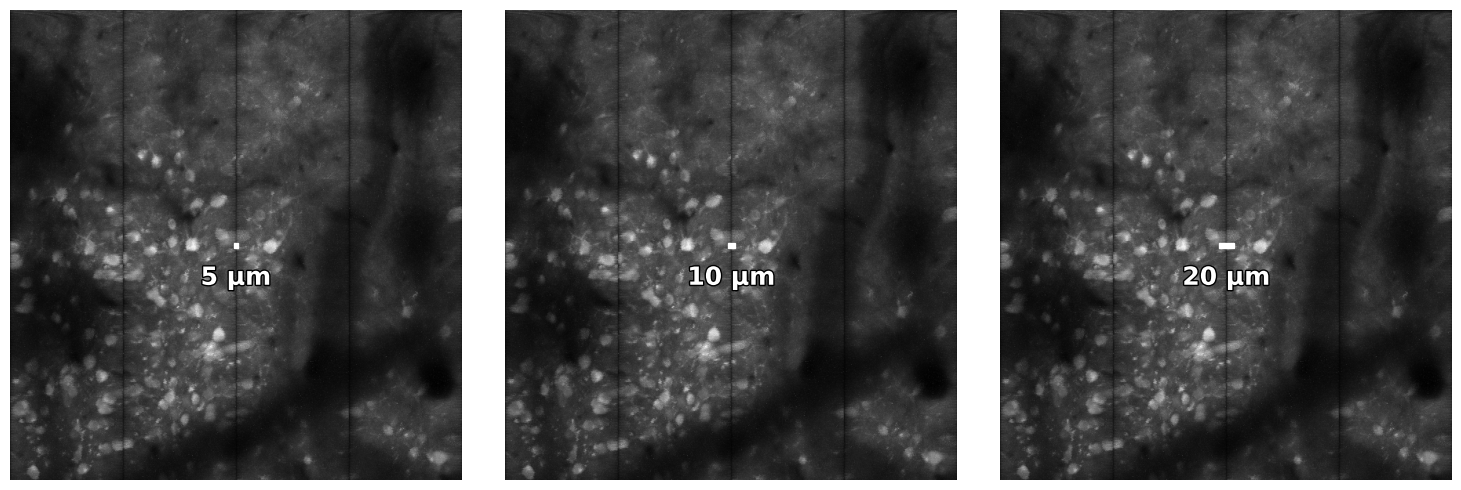
plot_data_with_scalebars will give you 3 images, at 5, 10 and 20 um.
You can use any summary images:
# a single frame
plot_with_scalebars(data[0, :, :], np.mean(dxy))
# mean projection image
plot_with_scalebars(np.mean(data, axis=0), np.mean(dxy))
# maximum projection image
plot_with_scalebars(np.max(data, axis=0), np.mean(dxy))
from lbm_caiman_python import plot_with_scalebars
# plot_with_scalebars(data[0, :, :], np.mean(dxy))
plot_with_scalebars(np.max(data, axis=0), np.mean(dxy))
3.2.2. Scaling patches relative to neuron size#
The generate_patch_view function divides the image into patches the same way CaImAn will do internally.
Increasing / decreasing the target_patch_size and overlap_fraction parameters to examine the effect of different stride/overlap values displayed in the title.
Tip
We want each patch to have “landmarks” (neurons) to use for alignment, so we want at least a few full neurons for each patch.
For this reason, we use the neuron_size * scale_factor as our target_patch_size.
We want to avoid neurons occupying the inner regions of multiple neighboring patches.
neuron_size = 15 # in micron
# - value of 1 makes patches ~neuron sized
# - value of 2 makes patches ~2x neuron sized
# - the larger your neurons, the larger you want to make this number
scale_factor = 3 # must be > 0
overlap_fraction = 0.3 # 30% of the patch size will be overlap
from lbm_caiman_python import generate_patch_view
target_patch_size=neuron_size*scale_factor
# this function assumes a square pixel resolution, so take the mean
fig, ax, stride_px, overlap_px = generate_patch_view(np.max(data, axis=0), pixel_resolution=np.mean(dxy), target_patch_size=target_patch_size, overlap_fraction=overlap_fraction)
plt.show()
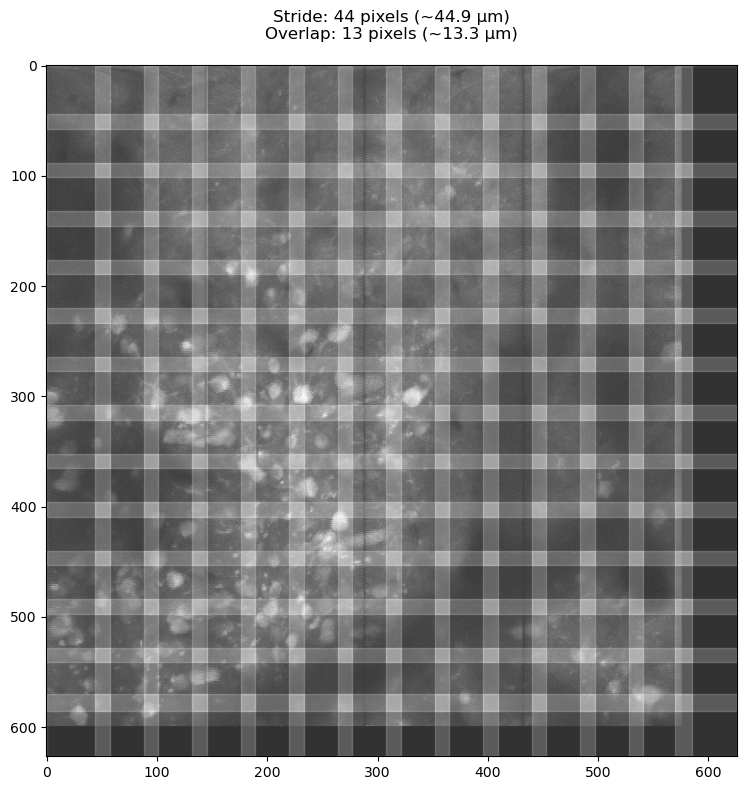
print(f'stride : {stride_px}px\noverlap: {overlap_px}px')
stride : 44px
overlap: 13px
3.2.3. Parameters from metadata#
lcp.params_from_metadata(metadata) will calculate sensible defaults for registration (and segmentation).
Compare the strides / overlaps generated from the generate_patch_view() function. If they are significantly different,
we recommend you use the defaults for the first run, and tune with the values from your custom set overlap.
params = lcp.params_from_metadata(metadata)
params
Overlaps too small. Increasing to neuron diameter.
{'main': {'pw_rigid': True,
'max_shifts': [10, 10],
'strides': [62, 64],
'overlaps': [7, 7],
'min_mov': None,
'gSig_filt': [0, 0],
'max_deviation_rigid': 3,
'border_nan': 'copy',
'splits_els': 14,
'upsample_factor_grid': 4,
'use_cuda': False,
'num_frames_split': 50,
'niter_rig': 1,
'is3D': False,
'splits_rig': 14,
'num_splits_to_process_rig': None,
'fr': 9.60806,
'dxy': [1.04, 1.0],
'decay_time': 0.4,
'p': 2,
'nb': 3,
'K': 20,
'rf': 34,
'stride': 7,
'gSig': 7.5,
'gSiz': (31.0, 31.0),
'method_init': 'greedy_roi',
'rolling_sum': True,
'use_cnn': False,
'ssub': 1,
'tsub': 1,
'merge_thr': 0.7,
'bas_nonneg': True,
'min_SNR': 1.4,
'rval_thr': 0.8},
'refit': True}
3.2.4. Tune gSig_filt (high-pass-filter)#
Use a spatial filter to highlight cells and eliminate low-frequency spatial information (i.e. blood-vessels)
In the below widget, move the slider at the bottom titled ‘gSig_filt’ to see if adding a gaussian blur can help highlight neurons and hide blood-vessels.
from ipywidgets import IntSlider, VBox
from caiman.motion_correction import high_pass_filter_space
slider_gsig_filt = IntSlider(value=1, min=1, max=33, step=1, description="high-pass-filter (σ)")
funcs = {1: lambda frame: high_pass_filter_space(frame, (slider_gsig_filt.value, slider_gsig_filt.value))}
# input movie will be shown on left, filtered on right
iw_gs = fpl.ImageWidget(
data=[data, data],
frame_apply=funcs,
names=["raw", f"filtered"],
figure_kwargs={"size": (1200, 600)},
cmap="gnuplot2"
)
iw_gs.figure["filtered"].set_title(f"filtered σ={slider_gsig_filt.value}")
def force_update(*args):
# forces the images to update when the gSig_filt slider is moved
iw_gs.current_index = iw_gs.current_index
iw_gs.reset_vmin_vmax()
iw_gs.figure["filtered"].set_title(f"filtered σ={slider_gsig_filt.value}")
iw_gs.reset_vmin_vmax()
slider_gsig_filt.observe(force_update, "value")
VBox([iw_gs.show(), slider_gsig_filt])
iw_gs.close(); slider_gsig_filt.close()
3.2.5. Change a parameter#
params['main']['gSig_filt'] = (2, 2)
3.3. Single-plane registration#
See the mesmerize-core utility docs for more information on batch creation.
We simple need to:
Add the batch item
Run the batch item
3.3.1. Add an item to the batch#
A “batch item” consists of:
algorithm to run,
algocurrently: mcorr, cnmf, cnmfe
input movie to run the algorithm on,
input_movie_pathcan be string or dataframe row
parameters for the specified algorithm,
paramsa name for you to keep track of things
item_namecan be anything
df.caiman.add_item(
algo='mcorr',
input_movie_path=file,
params=params,
item_name='mcorr',
)
df
| algo | item_name | input_movie_path | params | outputs | added_time | ran_time | algo_duration | comments | uuid | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': [10, 10], 'strides': [62, 64], 'overlaps': [7, 7], 'min_mov': None, 'gSig_... | None | 2025-01-23T22:01:50 | None | None | None | 22323919-7768-4d39-88ad-b0c3afff912f |
3.3.2. Run the batch item#
Note
On Linux & Mac it will run in subprocess but on Windows it will run in the local kernel. For this reason, on windows you need to reload the dataframe:
df=df.caiman.reload_from_disk()
df
If you ever get errors like
TypeError: NoneType is not subscriptable
This likely means you need to reload the dataframe.
Warning
On windows, df.iloc[i].caiman.run() will sometimes stall if you run additional cells before it completes.
df.iloc[0].caiman.run()
Running 22323919-7768-4d39-88ad-b0c3afff912f with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
<lbm_mc.caiman_extensions.common.DummyProcess at 0x2924802f490>
df=df.caiman.reload_from_disk()
df
| algo | item_name | input_movie_path | params | outputs | added_time | ran_time | algo_duration | comments | uuid | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': [10, 10], 'strides': [62, 64], 'overlaps': [7, 7], 'min_mov': None, 'gSig_... | {'mean-projection-path': '22323919-7768-4d39-88ad-b0c3afff912f/22323919-7768-4d39-88ad-b0c3afff912f_mean_projection.... | 2025-01-23T22:01:50 | 2025-01-23T22:03:07 | 71.93 sec | None | 22323919-7768-4d39-88ad-b0c3afff912f |
3.3.3. Check for errors in outputs#
In the table header outputs, you should see
{'mean-projection-path': ...}
If you see instead:
{'success': False, ...}
Run the below cell to evaluate the error message.
import pprint
pprint.pprint(df.iloc[0].outputs["traceback"])
None
3.3.4. Preview results: side-by-side comparison#
mesmerize-corelets us easily retrieve a memory-mappable moviefastplotlibwill efficiently display this movie using your GPU
See the mesmerize-core motion-correction (mcorr) API.
# get the motion corrected movie memmap
mcorr_movie = df.iloc[0].mcorr.get_output()
# the input movie, note that we use `.caiman` here instead of `.mcorr`
input_movie = df.iloc[0].caiman.get_input_movie()
Its helpful to add a mean window and zoom into a cell to see if movement was corrected.
mcorr_iw = fpl.ImageWidget(
data=[input_movie, mcorr_movie],
names=['raw', 'registered'],
cmap="gnuplot2",
window_funcs={"t": (np.mean, 17)}, # window functions, can change to np.max, np.std, anything that operates on a timeseries
figure_kwargs={"size": (900, 560)},
histogram_widget=False, # helps keep plots close together
)
mcorr_iw.show()
mcorr_iw.close()
3.4. Grid-search for best parameters#
3.4.1. scale_factor and gSig_filt#
If you still see non-rigid motion in your movie, we can try increasing the scale_factor to increase our patch-size or decrease it.
More than likely you will want to decrease the scale factor to process more patches.
The patched graphs displayed show you what patches each of your movie will use.
Tip
The item_name parameter of df.caiman.add_item() is used to group similar batch results.
You should keep the item_name the same for grid search items.
from copy import deepcopy
for scale in (1, 2, 4): # we started with a scale of 3
for gSig in (0, 1, 2, 4): # very exaustive
target_patch_size=neuron_size*scale
overlap_fraction = .3 # keep overlap to 30% of patch size
# _, _, stride_px, overlap_px = generate_patch_view(np.max(data, axis=0), pixel_resolution=np.mean(dxy), target_patch_size=target_patch_size, overlap_fraction=overlap_fraction)
stride = int(target_patch_size / pixel_resolution)
overlap = int(overlap_fraction * stride)
# deep copy is the safest way to copy dicts
new_params = deepcopy(params)
new_shift = int(np.ceil(overlap_px / 2))
new_stride = int(np.ceil(stride_px))
new_overlap = int(np.ceil(overlap_px))
new_params["main"]["max_shifts"] = (new_shift, new_shift)
new_params["main"]["strides"] = (new_stride, new_stride)
new_params["main"]["overlaps"] = (new_overlap, new_overlap)
new_params["main"]["overlaps"] = (new_overlap, new_overlap)
new_params["main"]["gSig_filt"] = (gSig, gSig)
df.caiman.add_item(
algo='mcorr',
input_movie_path=file,
params=new_params,
item_name='mcorr',
)
3.4.2. View the parameter differences#
Tip
We can use the caiman.get_params_diffs() to see the unique parameters between rows with the same item_name.
This shows the parameters that differ between our batch items.
diffs = df.caiman.get_params_diffs(algo="mcorr", item_name=df.iloc[0]["item_name"])
diffs
| max_shifts | overlaps | strides | gSig_filt | |
|---|---|---|---|---|
| 0 | [10, 10] | [7, 7] | [62, 64] | [0, 0] |
| 1 | (2, 2) | (4, 4) | (14, 14) | (0, 0) |
| 2 | (2, 2) | (4, 4) | (14, 14) | (1, 1) |
| 3 | (2, 2) | (4, 4) | (14, 14) | (2, 2) |
| 4 | (2, 2) | (4, 4) | (14, 14) | (4, 4) |
| 5 | (4, 4) | (8, 8) | (29, 29) | (0, 0) |
| 6 | (4, 4) | (8, 8) | (29, 29) | (1, 1) |
| 7 | (4, 4) | (8, 8) | (29, 29) | (2, 2) |
| 8 | (4, 4) | (8, 8) | (29, 29) | (4, 4) |
| 9 | (9, 9) | (17, 17) | (58, 58) | (0, 0) |
| 10 | (9, 9) | (17, 17) | (58, 58) | (1, 1) |
| 11 | (9, 9) | (17, 17) | (58, 58) | (2, 2) |
| 12 | (9, 9) | (17, 17) | (58, 58) | (4, 4) |
3.4.3. Use can also your own grid-search values.#
For example:
from copy import deepcopy
for shifts in [2,32]:
for strides in [12,64]:
overlaps = int(strides / 2)
# deep copy is the safest way to copy dicts
new_params = deepcopy(mcorr_params)
# assign the "max_shifts"
new_params["main"]["pw_rigid"] = True
new_params["main"]["max_shifts"] = (shifts, shifts)
new_params["main"]["strides"] = (strides, strides)
new_params["main"]["overlaps"] = (overlaps, overlaps)
df.caiman.add_item(
algo='mcorr',
input_movie_path=file,
params=new_params,
item_name='mcorr', # filename of the movie, but can be anything
)
df.caiman.reload_from_disk()
3.4.4. Run all parameter sets#
df.iterrows() iterates through rows and returns the numerical index and row for each iteration
for i, row in df.iterrows():
if row["outputs"] is not None: # item has already been run
continue # skip
process = row.caiman.run()
# on Windows you MUST reload the batch dataframe after every iteration because it uses the `local` backend.
# this is unnecessary on Linux & Mac
# "DummyProcess" is used for local backend so this is automatic
if process.__class__.__name__ == "DummyProcess":
df = df.caiman.reload_from_disk()
Running 8d6ff88a-12d4-4029-8b41-064b13d5ff43 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 958a6e01-8fb3-4ae5-b06b-623bce6782ae with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 58cd4cb9-e571-49a0-9c73-a33e6fdb83e2 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 1842685d-e421-423b-b872-3c76cb684fa4 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running f8d5343d-14a3-4872-89d6-6f862f9ce255 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 9ad664bf-c22e-4ba6-ac2d-a7605933e2d2 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 90963ca2-a226-4503-8781-33fe09b40960 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 329e34f0-3c45-4427-820a-8000fc57c054 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
3.4.5. Check for errors#
# make sure everything works properly
# check for outputs {'success': False, ...
df = df.caiman.reload_from_disk()
df.outputs
0 {'mean-projection-path': '22323919-7768-4d39-88ad-b0c3afff912f/22323919-7768-4d39-88ad-b0c3afff912f_mean_projection....
1 {'mean-projection-path': 'efd3fcee-ba9e-46ff-826e-cffbef906411/efd3fcee-ba9e-46ff-826e-cffbef906411_mean_projection....
2 {'mean-projection-path': 'ab1e18e8-d860-408c-8588-a7995f542639/ab1e18e8-d860-408c-8588-a7995f542639_mean_projection....
3 {'mean-projection-path': '8d6ff88a-12d4-4029-8b41-064b13d5ff43/8d6ff88a-12d4-4029-8b41-064b13d5ff43_mean_projection....
4 {'mean-projection-path': '958a6e01-8fb3-4ae5-b06b-623bce6782ae/958a6e01-8fb3-4ae5-b06b-623bce6782ae_mean_projection....
5 {'mean-projection-path': '58cd4cb9-e571-49a0-9c73-a33e6fdb83e2/58cd4cb9-e571-49a0-9c73-a33e6fdb83e2_mean_projection....
6 {'mean-projection-path': '1842685d-e421-423b-b872-3c76cb684fa4/1842685d-e421-423b-b872-3c76cb684fa4_mean_projection....
7 {'mean-projection-path': 'f8d5343d-14a3-4872-89d6-6f862f9ce255/f8d5343d-14a3-4872-89d6-6f862f9ce255_mean_projection....
8 {'mean-projection-path': 'd6ccef75-5a5f-4d30-8dfa-2e1fe493fee1/d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1_mean_projection....
9 {'mean-projection-path': '9ad664bf-c22e-4ba6-ac2d-a7605933e2d2/9ad664bf-c22e-4ba6-ac2d-a7605933e2d2_mean_projection....
10 {'mean-projection-path': '90963ca2-a226-4503-8781-33fe09b40960/90963ca2-a226-4503-8781-33fe09b40960_mean_projection....
11 {'mean-projection-path': '19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e/19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e_mean_projection....
12 {'mean-projection-path': '329e34f0-3c45-4427-820a-8000fc57c054/329e34f0-3c45-4427-820a-8000fc57c054_mean_projection....
Name: outputs, dtype: object
3.5. Evaluate results#
3.5.1. Summary statistics#
You can evaluate some basic statistics like mean, median, and percentiles of each item in your batch results.
It is possible that parameter sets you used for your grid-search yield the same output movie (i.e. overlaps of (2, 2) and (3, 3) likely yield the same result).
You will want to compute registration statistics (correlation, optical flow) later which is computationally expensive, so it helps to limit the batch items you will use as input.
summary_df = lcp.get_summary_batch(df)
summary_df.head()
| algo | Runs | Successful | Unsuccessful | |
|---|---|---|---|---|
| 0 | mcorr | 13 | 13 | 0 |
| 1 | cnmf | 0 | 0 | 0 |
Find the duplicates#
Before calculating computationally intensive summary registration statistics like correlation and flow, we want to remove duplicates to avoid unnessesary computaion.
lcp.drop_duplicates will iterate over your dataframe and drop any duplicate results.
# make sure to use the batch dataframe, not the summary dataframe
df = lcp.drop_duplicates(df)
Removing duplicate item efd3fcee-ba9e-46ff-826e-cffbef906411.
Removing duplicate item 58cd4cb9-e571-49a0-9c73-a33e6fdb83e2.
Removing duplicate item 9ad664bf-c22e-4ba6-ac2d-a7605933e2d2.
Reload our dataframe if you made a mistake#
df = df.caiman.reload_from_disk()
df.head()
| algo | item_name | input_movie_path | params | outputs | added_time | ran_time | algo_duration | comments | uuid | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': [10, 10], 'strides': [62, 64], 'overlaps': [7, 7], 'min_mov': None, 'gSig_... | {'mean-projection-path': '22323919-7768-4d39-88ad-b0c3afff912f/22323919-7768-4d39-88ad-b0c3afff912f_mean_projection.... | 2025-01-23T22:01:50 | 2025-01-23T22:03:07 | 71.93 sec | None | 22323919-7768-4d39-88ad-b0c3afff912f |
| 1 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | {'mean-projection-path': 'ab1e18e8-d860-408c-8588-a7995f542639/ab1e18e8-d860-408c-8588-a7995f542639_mean_projection.... | 2025-01-23T23:46:33 | 2025-01-23T23:51:40 | 126.03 sec | None | ab1e18e8-d860-408c-8588-a7995f542639 |
| 2 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | {'mean-projection-path': '8d6ff88a-12d4-4029-8b41-064b13d5ff43/8d6ff88a-12d4-4029-8b41-064b13d5ff43_mean_projection.... | 2025-01-23T23:46:34 | 2025-01-24T10:34:48 | 124.27 sec | None | 8d6ff88a-12d4-4029-8b41-064b13d5ff43 |
| 3 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | {'mean-projection-path': '958a6e01-8fb3-4ae5-b06b-623bce6782ae/958a6e01-8fb3-4ae5-b06b-623bce6782ae_mean_projection.... | 2025-01-23T23:46:34 | 2025-01-24T10:36:55 | 126.95 sec | None | 958a6e01-8fb3-4ae5-b06b-623bce6782ae |
| 4 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (4, 4), 'strides': (29, 29), 'overlaps': (8, 8), 'min_mov': None, 'gSig_fi... | {'mean-projection-path': '1842685d-e421-423b-b872-3c76cb684fa4/1842685d-e421-423b-b872-3c76cb684fa4_mean_projection.... | 2025-01-23T23:46:34 | 2025-01-24T10:39:52 | 89.0 sec | None | 1842685d-e421-423b-b872-3c76cb684fa4 |
3.5.2. Evaluation metrics#
Metric |
Description |
Higher or Lower is Better? |
|---|---|---|
Correlations |
How similar each frame is to a reference “template” frame. |
Higher is better (closer to 1 means frames are more similar) |
Norms |
The amount of movement or change in each frame. |
Lower is better (less movement indicates stability) |
Smoothness |
How smooth/sharp/crisp the image appears. |
Higher is better (crisper mean image) |
Flows |
The direction and speed of pixel movement between frames. |
Lower “better” (less movement relative to mean image) |
Smoothness (correlations) |
How smooth the image of frame-to-frame correlations appears. |
Higher is better (higher smoothness means sharper correlation map) |
Image Correlations |
Shows the local similarity of neighboring pixels in each frame. |
Higher is better (higher local similarity means more consistent images) |
Many of these metrics are related. Norms is a measure of the stregnth of flows (which also include direction). Smoothness measures sharpness of edges which typically result in highcorrelations because individual pixels are located in the same location.
For more a technical discussion on these metrics, see NoRMCorre’s companion paper (section 2.2.1 - 2.2.4, 3.2.1 - 3.2.4).
Compute batch metrics#
Feed your batch dataframe to compute_batch_metrics to compute each of these metrics and save them to disk.
This functiion will attempt to locate your raw data path to run evaluation metrics on the raw input-file for comparison.
Use overwrite=False to avoid re-calculating these values.
Warning
compute_batch_metrics() is a computationally intensive function. ~70s per batch item on a 600 x 576 pixel image with 1760 frames.
The returned metrics_files containes a list of paths to the saved metrics.
metrics_df = lcp.get_summary_mcorr(df)
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 214.51it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.93it/s]
Computed metrics for raw data in 60.47 seconds.
Processing batch index 0...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 210.83it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 9.03it/s]
Computed metrics for batch index 0 in 60.22 seconds.
Processing batch index 1...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 202.53it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.91it/s]
Computed metrics for batch index 1 in 61.36 seconds.
Processing batch index 2...
Correlations: 100%|██████████████████████████| 1729/1729 [00:07<00:00, 217.11it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.89it/s]
Computed metrics for batch index 2 in 60.87 seconds.
Processing batch index 3...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 211.99it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.98it/s]
Computed metrics for batch index 3 in 60.46 seconds.
Processing batch index 4...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 209.26it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 9.02it/s]
Computed metrics for batch index 4 in 60.60 seconds.
Processing batch index 5...
Correlations: 100%|██████████████████████████| 1729/1729 [00:07<00:00, 218.73it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.93it/s]
Computed metrics for batch index 5 in 60.30 seconds.
Processing batch index 6...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 206.74it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.99it/s]
Computed metrics for batch index 6 in 60.51 seconds.
Processing batch index 7...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 212.33it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 9.01it/s]
Computed metrics for batch index 7 in 60.42 seconds.
Processing batch index 8...
Correlations: 100%|██████████████████████████| 1729/1729 [00:08<00:00, 209.08it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 9.01it/s]
Computed metrics for batch index 8 in 60.29 seconds.
Processing batch index 9...
Correlations: 100%|██████████████████████████| 1729/1729 [00:07<00:00, 218.50it/s]
Optical flow: 100%|█████████████████████████████| 345/345 [00:38<00:00, 8.92it/s]
Computed metrics for batch index 9 in 60.55 seconds.
View numerical results#
lcp.get_summary_mcorr() will use the resulting filepaths to load the mean metric statistic, along with the path to the metrics file.
This gives you an overview of the best results. The metric_path is needed to load larger arrays for plotting that don’t performantly fit into a dataframe cell.
Note
batch_index = -1 for your raw data. This does not correspond to the ‘last index’, as it does with numpy slicing. It is just convenient to avoid casting another data type like str/np.null.
metrics_df
| mean_corr | mean_norm | crispness | uuid | batch_index | metric_path | |
|---|---|---|---|---|---|---|
| 0 | 0.889579 | 264.345490 | 19.149659 | raw_628bcc12-4d11-4c1f-aa64-cb47526867f4 | -1 | C:\Users\RBO\lbm_data\processed\plane_11_assembled_metrics.npz |
| 1 | 0.917335 | 266.898865 | 19.722043 | 22323919-7768-4d39-88ad-b0c3afff912f | 0 | C:\Users\RBO\lbm_data\processed\batch\22323919-7768-4d39-88ad-b0c3afff912f\22323919-7768-4d39-88ad-b0c3afff912f-plan... |
| 2 | 0.903623 | 151.739716 | 19.741791 | ab1e18e8-d860-408c-8588-a7995f542639 | 1 | C:\Users\RBO\lbm_data\processed\batch\ab1e18e8-d860-408c-8588-a7995f542639\ab1e18e8-d860-408c-8588-a7995f542639-plan... |
| 3 | 0.906983 | 68.337738 | 20.577532 | 8d6ff88a-12d4-4029-8b41-064b13d5ff43 | 2 | C:\Users\RBO\lbm_data\processed\batch\8d6ff88a-12d4-4029-8b41-064b13d5ff43\8d6ff88a-12d4-4029-8b41-064b13d5ff43-plan... |
| 4 | 0.907472 | 105.332611 | 20.506616 | 958a6e01-8fb3-4ae5-b06b-623bce6782ae | 3 | C:\Users\RBO\lbm_data\processed\batch\958a6e01-8fb3-4ae5-b06b-623bce6782ae\958a6e01-8fb3-4ae5-b06b-623bce6782ae-plan... |
| 5 | 0.899842 | 180.179504 | 19.350287 | 1842685d-e421-423b-b872-3c76cb684fa4 | 4 | C:\Users\RBO\lbm_data\processed\batch\1842685d-e421-423b-b872-3c76cb684fa4\1842685d-e421-423b-b872-3c76cb684fa4-plan... |
| 6 | 0.903309 | 88.548859 | 20.409476 | f8d5343d-14a3-4872-89d6-6f862f9ce255 | 5 | C:\Users\RBO\lbm_data\processed\batch\f8d5343d-14a3-4872-89d6-6f862f9ce255\f8d5343d-14a3-4872-89d6-6f862f9ce255-plan... |
| 7 | 0.905676 | 85.730988 | 20.597341 | d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1 | 6 | C:\Users\RBO\lbm_data\processed\batch\d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1\d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1-plan... |
| 8 | 0.897103 | 206.667816 | 19.136277 | 90963ca2-a226-4503-8781-33fe09b40960 | 7 | C:\Users\RBO\lbm_data\processed\batch\90963ca2-a226-4503-8781-33fe09b40960\90963ca2-a226-4503-8781-33fe09b40960-plan... |
| 9 | 0.902426 | 105.812897 | 20.490284 | 19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e | 8 | C:\Users\RBO\lbm_data\processed\batch\19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e\19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e-plan... |
| 10 | 0.905927 | 80.540215 | 20.706461 | 329e34f0-3c45-4427-820a-8000fc57c054 | 9 | C:\Users\RBO\lbm_data\processed\batch\329e34f0-3c45-4427-820a-8000fc57c054\329e34f0-3c45-4427-820a-8000fc57c054-plan... |
Merge metrics results with added parameters used for grid-search#
We used df.caiman.get_param_diffs() to get a list of parameters that differ between batch runs, to filter out the parameters common to each item.
We can add these values as columns to evaluate which parameters led to the desired metric by using add_param_diffs with either or both of the result tables we just calculated.
param_diffs = df.caiman.get_params_diffs("mcorr", item_name=df.iloc[0]["item_name"])
results = lcp.concat_param_diffs(metrics_df, param_diffs)
results
# results.sort_values('mean_norm', ascending=True)
| mean_corr | mean_norm | crispness | gSig_filt | strides | overlaps | max_shifts | batch_index | uuid | metric_path | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.889579 | 264.345490 | 19.149659 | None | None | None | None | -1 | raw_628bcc12-4d11-4c1f-aa64-cb47526867f4 | C:\Users\RBO\lbm_data\processed\plane_11_assembled_metrics.npz |
| 1 | 0.917335 | 266.898865 | 19.722043 | [0, 0] | [62, 64] | [7, 7] | [10, 10] | 0 | 22323919-7768-4d39-88ad-b0c3afff912f | C:\Users\RBO\lbm_data\processed\batch\22323919-7768-4d39-88ad-b0c3afff912f\22323919-7768-4d39-88ad-b0c3afff912f-plan... |
| 2 | 0.903623 | 151.739716 | 19.741791 | (1, 1) | (14, 14) | (4, 4) | (2, 2) | 1 | ab1e18e8-d860-408c-8588-a7995f542639 | C:\Users\RBO\lbm_data\processed\batch\ab1e18e8-d860-408c-8588-a7995f542639\ab1e18e8-d860-408c-8588-a7995f542639-plan... |
| 3 | 0.906983 | 68.337738 | 20.577532 | (2, 2) | (14, 14) | (4, 4) | (2, 2) | 2 | 8d6ff88a-12d4-4029-8b41-064b13d5ff43 | C:\Users\RBO\lbm_data\processed\batch\8d6ff88a-12d4-4029-8b41-064b13d5ff43\8d6ff88a-12d4-4029-8b41-064b13d5ff43-plan... |
| 4 | 0.907472 | 105.332611 | 20.506616 | (4, 4) | (14, 14) | (4, 4) | (2, 2) | 3 | 958a6e01-8fb3-4ae5-b06b-623bce6782ae | C:\Users\RBO\lbm_data\processed\batch\958a6e01-8fb3-4ae5-b06b-623bce6782ae\958a6e01-8fb3-4ae5-b06b-623bce6782ae-plan... |
| 5 | 0.899842 | 180.179504 | 19.350287 | (1, 1) | (29, 29) | (8, 8) | (4, 4) | 4 | 1842685d-e421-423b-b872-3c76cb684fa4 | C:\Users\RBO\lbm_data\processed\batch\1842685d-e421-423b-b872-3c76cb684fa4\1842685d-e421-423b-b872-3c76cb684fa4-plan... |
| 6 | 0.903309 | 88.548859 | 20.409476 | (2, 2) | (29, 29) | (8, 8) | (4, 4) | 5 | f8d5343d-14a3-4872-89d6-6f862f9ce255 | C:\Users\RBO\lbm_data\processed\batch\f8d5343d-14a3-4872-89d6-6f862f9ce255\f8d5343d-14a3-4872-89d6-6f862f9ce255-plan... |
| 7 | 0.905676 | 85.730988 | 20.597341 | (4, 4) | (29, 29) | (8, 8) | (4, 4) | 6 | d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1 | C:\Users\RBO\lbm_data\processed\batch\d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1\d6ccef75-5a5f-4d30-8dfa-2e1fe493fee1-plan... |
| 8 | 0.897103 | 206.667816 | 19.136277 | (1, 1) | (58, 58) | (17, 17) | (9, 9) | 7 | 90963ca2-a226-4503-8781-33fe09b40960 | C:\Users\RBO\lbm_data\processed\batch\90963ca2-a226-4503-8781-33fe09b40960\90963ca2-a226-4503-8781-33fe09b40960-plan... |
| 9 | 0.902426 | 105.812897 | 20.490284 | (2, 2) | (58, 58) | (17, 17) | (9, 9) | 8 | 19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e | C:\Users\RBO\lbm_data\processed\batch\19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e\19d16022-e6e5-4bb6-b0fd-e9be9f7c4f9e-plan... |
| 10 | 0.905927 | 80.540215 | 20.706461 | (4, 4) | (58, 58) | (17, 17) | (9, 9) | 9 | 329e34f0-3c45-4427-820a-8000fc57c054 | C:\Users\RBO\lbm_data\processed\batch\329e34f0-3c45-4427-820a-8000fc57c054\329e34f0-3c45-4427-820a-8000fc57c054-plan... |
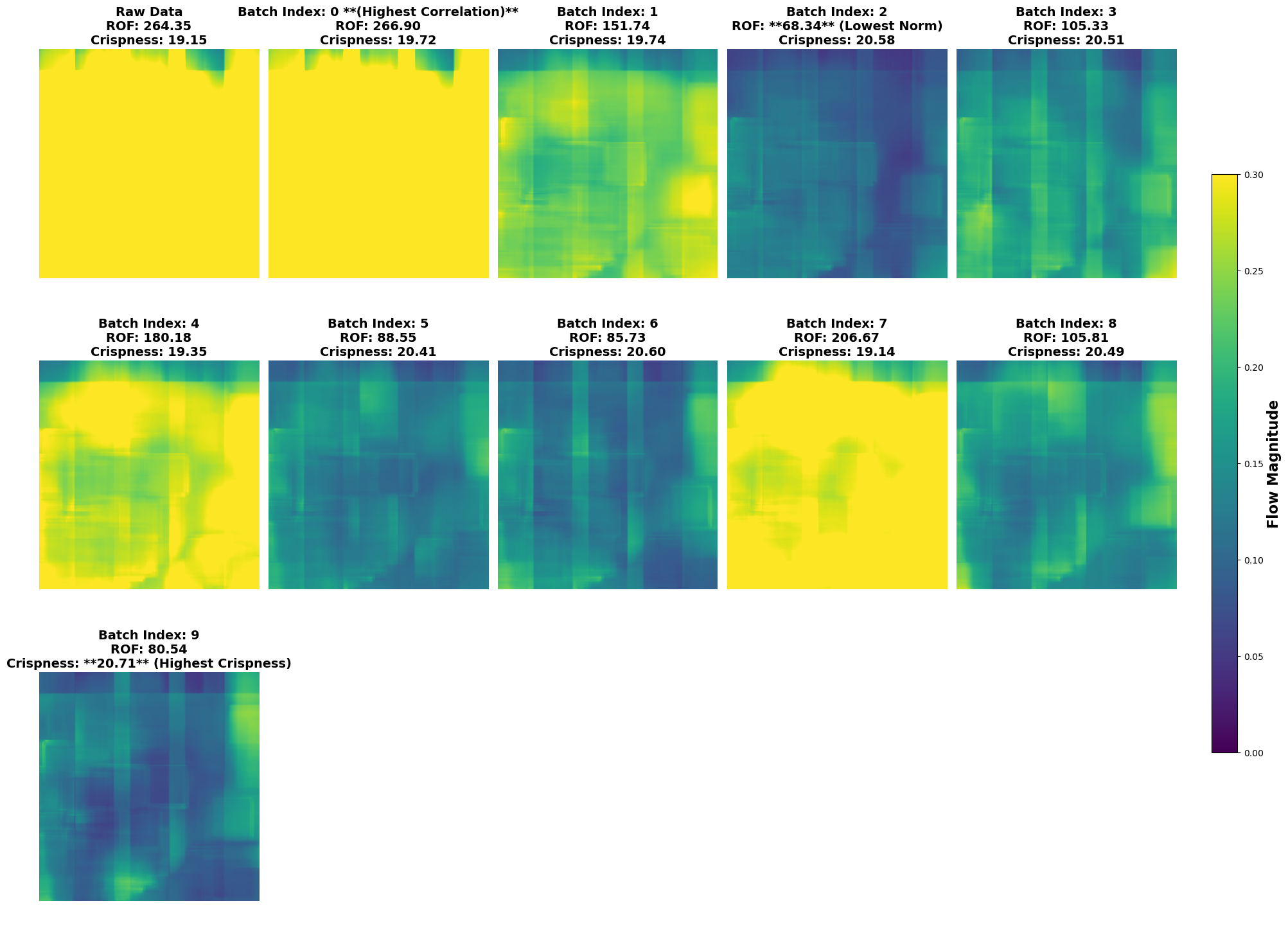
Optical Flow#
Optical flow measures the movement of a pixel between two consecutive frames.
Movement is illustrated as brighter values, with the hue (color) illustrating the direction of movement.
import lbm_caiman_python as lcp
# This will plot all batches.
lcp.plot_optical_flows(results, max_columns=5)
Plot residual flows and correlations#
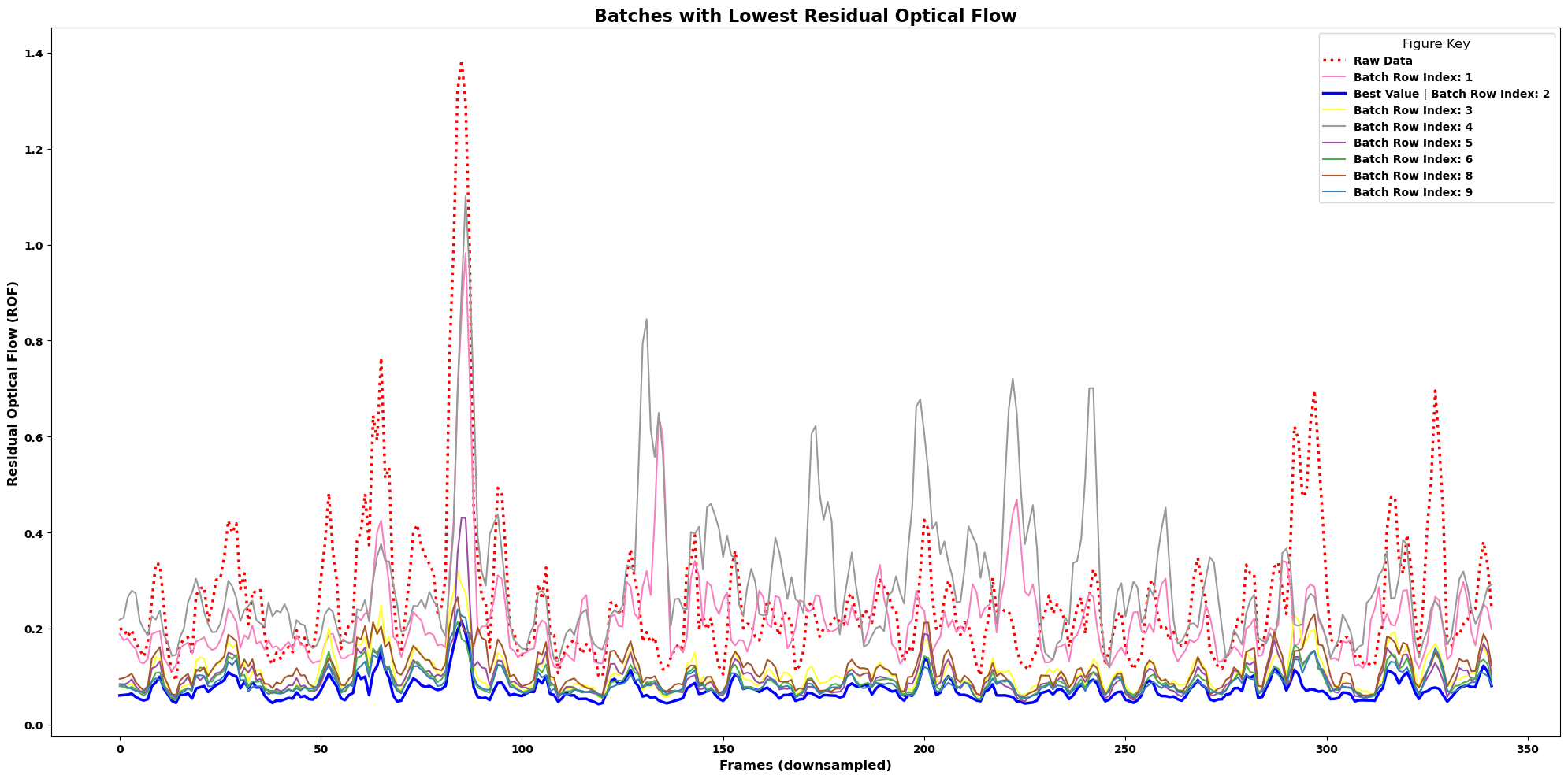
lcp.plot_residual_flows()andlcp.plot_correlations()will first sort your results by the corresponding metric.Use the
num_batchesargument to display just the best results and avoid cluttering your figureIncrease the number of batches to see groupings
You can then see the batch index in the key to query back to your metrics results and see if any parameters led to metrics being grouped
With many batches#
lcp.plot_residual_flows(results, num_batches=8, smooth=True, winsize=3)
With only 2 batches, no smoothing#
Now, show only the best 2 batch runs as determiend by residual optical flow without smoothing
Note
No matter how many batches you decide to include, the raw data will be displayed as well in a dotted line.
If your raw data is the best result, it will be a blue dotted line.
lcp.plot_residual_flows(results, num_batches=2, smooth=False)
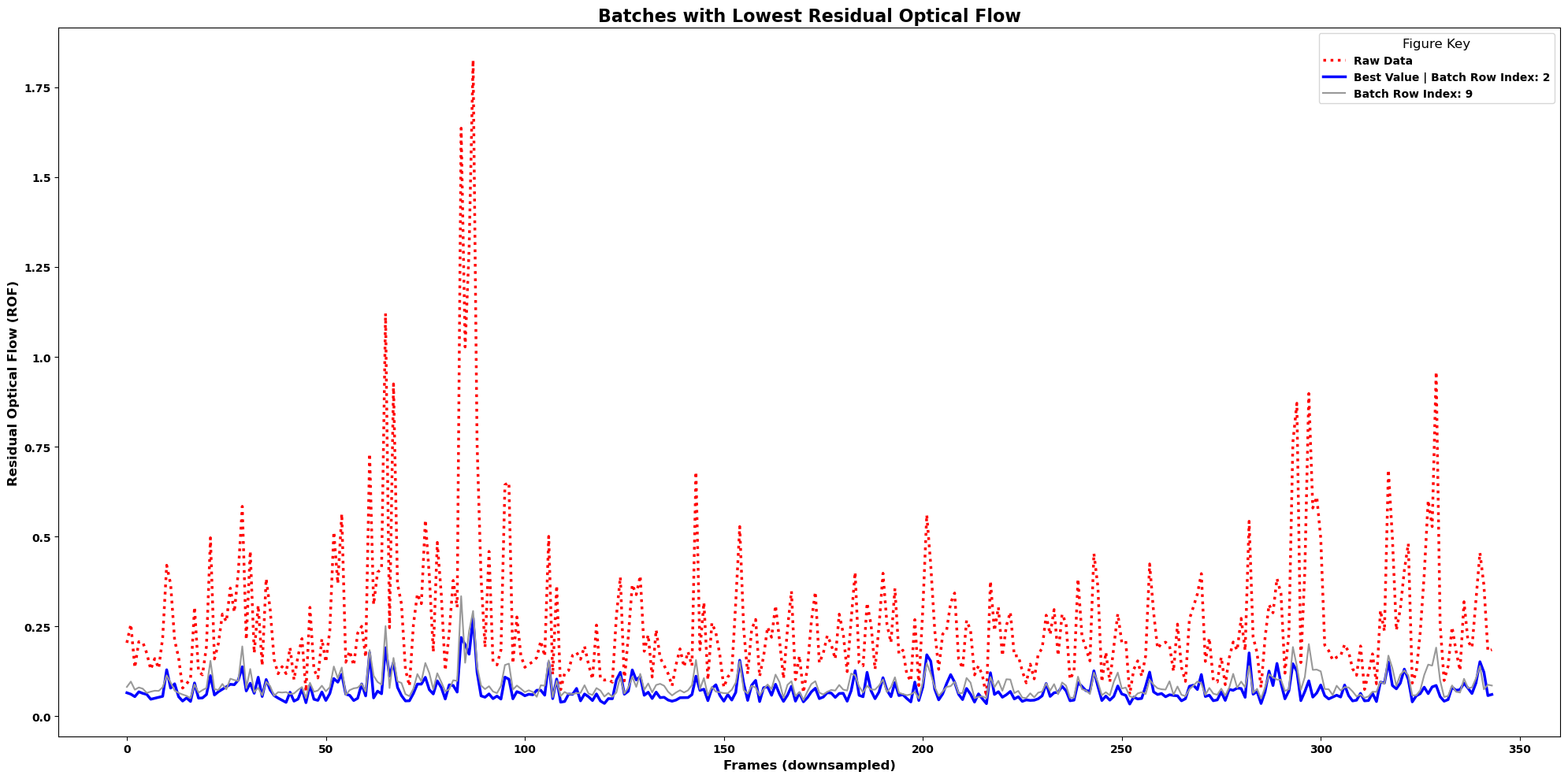
Notice how there are many batch results that seem to align perfectly with the “best” result.
Correlation here is defined as the mean pixel correlation for each frame to the correlation image within each movie.
It is possible to increase pixel-correlations and still have poor registration. Correlation and flow metrics should be used together.
lcp.plot_correlations(results, num_batches=8, smooth=True, winsize=10)
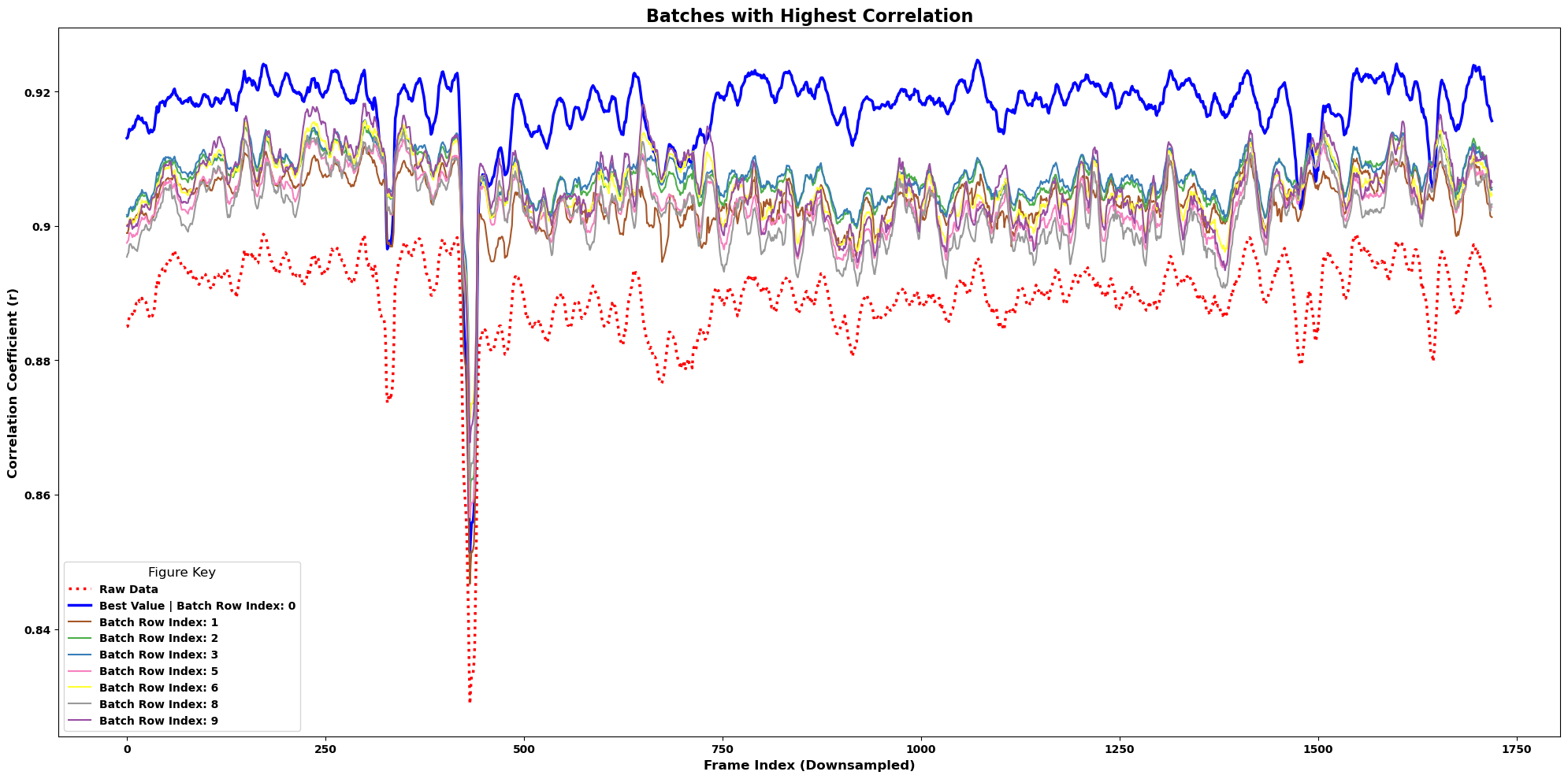
movie_A = df.iloc[0].mcorr.get_output() # highest correlation
movie_B = df.iloc[2].mcorr.get_output() # losest optical flow
# Change the size/shape depending on how many parameter sweep inputs you used
figure_kwargs={"size": (900, 700), "shape": (1, 2)}
mcorr_iw_multiple = fpl.ImageWidget(
data=[movie_A, movie_B], # list of movies
window_funcs={"t": (np.mean, 7)}, # window functions as a kwarg, this is what the slider was used for in the ready-made viz
figure_kwargs=figure_kwargs,
names=['Movie A', 'Movie B'], # subplot names used for titles
cmap="gnuplot2"
)
# free up some space
for subplot in mcorr_iw_multiple.figure:
subplot.docks["right"].size = 0
subplot.toolbar = False
mcorr_iw_multiple.show()
mcorr_iw_multiple.window_funcs["t"].window_size = 13
mcorr_iw_multiple.close()
3.6. Apply the ‘best parameters’ to remaining files#
When you decide which parameter set works the best, we keep it and delete the other batch items.
Remove batch items (i.e. rows) using df.caiman.remove_item(<item_uuid>). This also cleans up the output data in the batch directory.
Warning
On windows, you will get an error PermissionError: You do not have permissions to remove the output data for the batch item, aborting.
This can happen if you forgot to close one of the above widgets, or if you have a memory mapped file open.
There is currently no way to close a numpy.memmap: https://github.com/numpy/numpy/issues/13510
The solution is to restart your kernel. You will need to re-run the cells that define your batch/bath and reload your batch
with df = mc.load_batch(batch_path). Make sure mc.set_raw_parent_data_path() is in the re-run cells.
# UNCOMMENT THIS TO DELETE BATCH RUNS
# THIS IS COMMENTED OUT SO YOU DONT ACCIDENTALLY DELETE RESULTS YOU HAVE BEEN WAITING FOR ALL NIGHT!!!
rows_keep = [2]
for i, row in df.iterrows():
if i not in rows_keep:
df.caiman.remove_item(row.uuid, safe_removal=False)
df
| algo | item_name | input_movie_path | params | outputs | added_time | ran_time | algo_duration | comments | uuid | |
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | mcorr | mcorr | plane_11_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | {'mean-projection-path': '8d6ff88a-12d4-4029-8b41-064b13d5ff43/8d6ff88a-12d4-4029-8b41-064b13d5ff43_mean_projection.... | 2025-01-23T23:46:34 | 2025-01-24T10:34:48 | 124.27 sec | None | 8d6ff88a-12d4-4029-8b41-064b13d5ff43 |
for i in range(len(tiff_files)):
# don't re-process the same file
if tiff_files[i].name == df.iloc[0].input_movie_path:
continue
df.caiman.add_item(
algo='mcorr',
input_movie_path=tiff_files[i],
params= df.iloc[0].params, # use the same parameters
item_name=f'{tiff_files[i].name}', # filename of the movie, but can be anything
)
df.caiman.reload_from_disk()
df.tail()
| algo | item_name | input_movie_path | params | outputs | added_time | ran_time | algo_duration | comments | uuid | |
|---|---|---|---|---|---|---|---|---|---|---|
| 54 | mcorr | plane_5_assembled.tiff | plane_5_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | None | 2025-01-24T11:36:38 | None | None | None | 1f5f8773-da41-4475-aa46-3b92fd7bc2e6 |
| 55 | mcorr | plane_6_assembled.tiff | plane_6_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | None | 2025-01-24T11:36:38 | None | None | None | 0872f10e-8745-45bd-b2b3-6b8560f193a7 |
| 56 | mcorr | plane_7_assembled.tiff | plane_7_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | None | 2025-01-24T11:36:38 | None | None | None | 0374178f-22e0-4843-bcb9-5e6308b368b1 |
| 57 | mcorr | plane_8_assembled.tiff | plane_8_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | None | 2025-01-24T11:36:38 | None | None | None | d519cc68-a4ad-4fad-8c12-0cc2a1c4bc7a |
| 58 | mcorr | plane_9_assembled.tiff | plane_9_assembled.tiff | {'main': {'pw_rigid': True, 'max_shifts': (2, 2), 'strides': (14, 14), 'overlaps': (4, 4), 'min_mov': None, 'gSig_fi... | None | 2025-01-24T11:36:38 | None | None | None | 6f5d3d95-5cec-49aa-889a-d4c2194bcdb3 |
for i, row in df.iterrows():
if row["outputs"] is not None: # item has already been run
continue # skip
process = row.caiman.run()
# on Windows you MUST reload the batch dataframe after every iteration because it uses the `local` backend.
# this is unnecessary on Linux & Mac
# "DummyProcess" is used for local backend so this is automatic
if process.__class__.__name__ == "DummyProcess":
df = df.caiman.reload_from_disk()
Running cbdd2903-c3c3-495c-842a-a611cc536ab9 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running e60d2fd3-d8cc-44f3-87fb-bb14ecd661e0 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running f8469e8d-f381-4347-a807-3ffc5e2da32f with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running b42de600-4e8d-4c4c-bfae-633051431809 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 7774bb43-ad40-4240-867a-2c81a0e99eb7 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 9ad445d9-3e08-4547-b61d-c4ebeecff095 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 1777e77c-0529-459c-86a1-64d3bdc01584 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running c57ce669-9380-4c4d-b161-576f970b5b57 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running 6df4fcf2-0375-4027-9e9f-bb0a13247877 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
mc finished successfully!
computing projections
Computing correlation image
finished computing correlation image
Running e37174be-d7d9-4b0e-b438-61f080eaf691 with local backend
starting mc
The local backend is an alias for the multiprocessing backend, and the alias may be removed in some future version of Caiman
In setting CNMFParams, non-pathed parameters were used; this is deprecated. In some future version of Caiman, allow_legacy will default to False (and eventually will be removed)
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.
Movie average is negative. Removing 1st percentile.