4. Axial Collation and Correction#
Use pollen calibration data to spatially align each z-plane and merge across planes.
Core function(s): collatePlanes()
Note
Before the session being processed, you should have collected a cailbration file (i.e. pollen_calibration_00001.tif}).
4.1. Inputs#
Important
These files hold data used to align each z-plane depth around the same [Y, X] coordinates.
Place these files in the same directory as your segmentation_plane_N files.
First, the [Y, X] offsets (in microns) are used for an initial, dirty axial alignment:
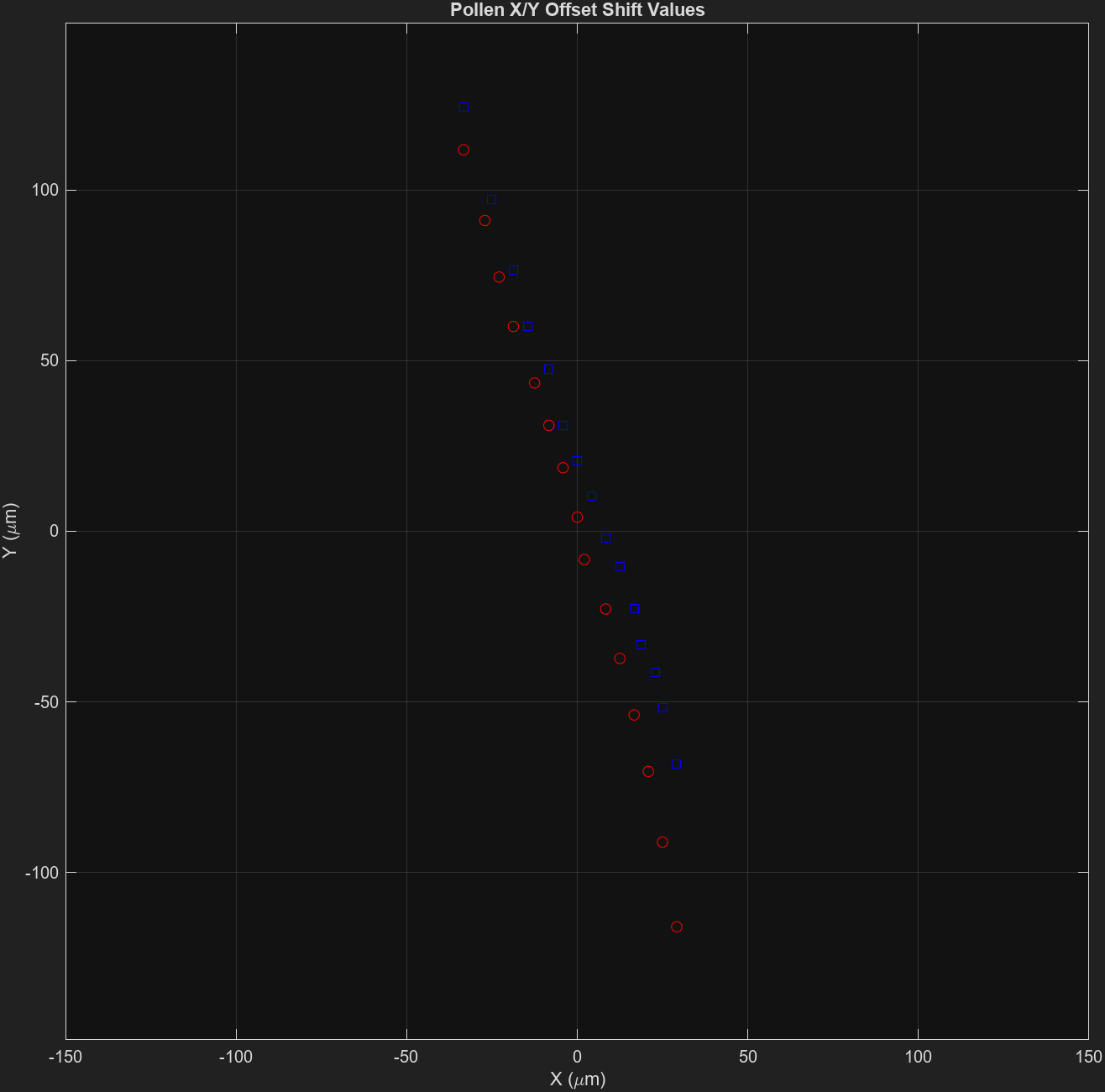
This alignment should improve the spatial consistency between z-planes, but there is a further refinement step which prompts a graphical interface for \(z-plane(n)\) and \(z-plane(n+1)\).
You will be prompted to select the same feature / region-of-interest / neuron.
After selecting 3 neurons for each plane, you are done with this LBM pipeline.
Depending on your axial field-of-view, there is likely neuronal contamination between z-planes. We can use this to select \(feature(z)\) in the left box and $feature(z+1) on the right box.
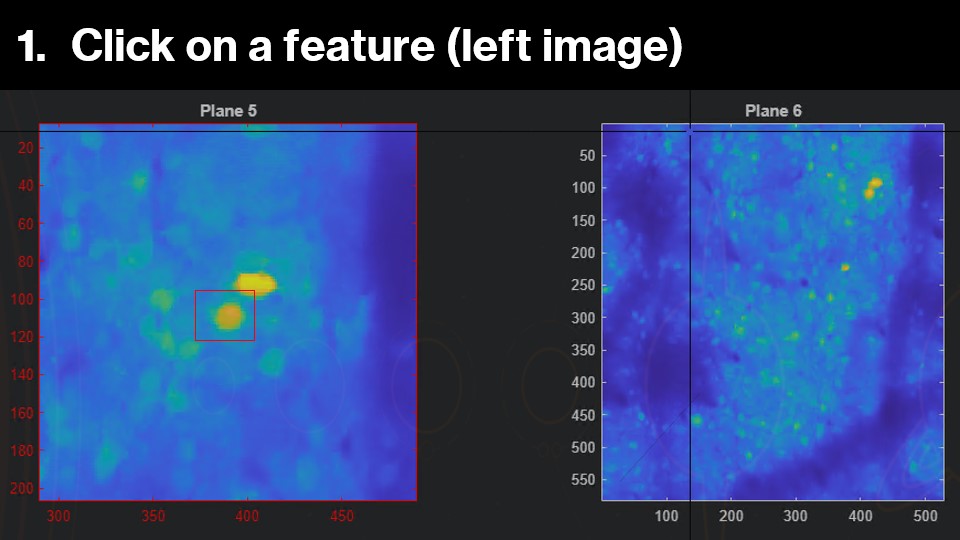
The image you should be clicking on will be highighted red. Once the selection is made, the right image will now zoom into the corresponding region in the next z-plane. Now, you can select the same feature in this plane:
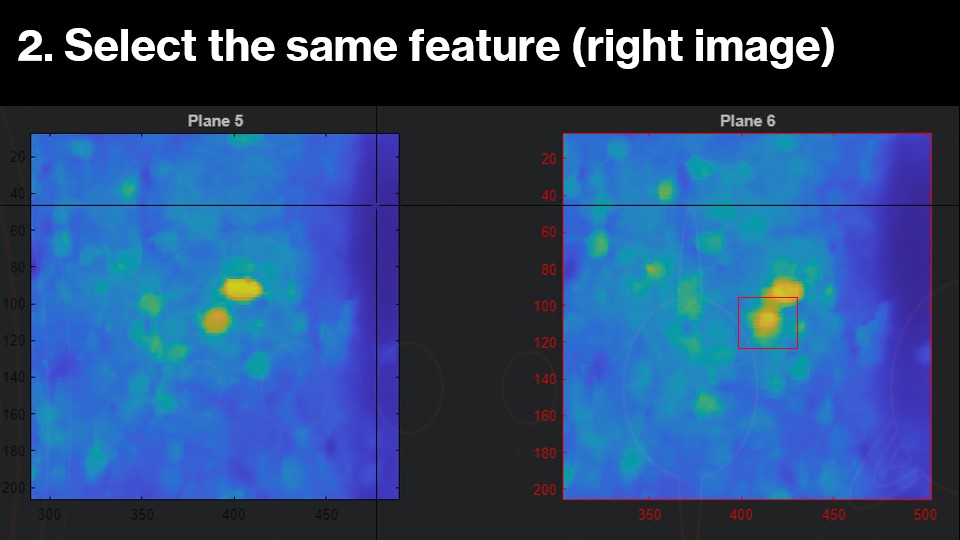
This makes a few assumptions about the axial distance between z-planes. First, each z-plane should be close enough in distance (for example, ~16um) that neuronal features will be similar and can be used for alignment.
4.2. Outputs#
.figfiles showing neuron distributions in z and radial directions.A
.matfile:caiman_collated_output_plane_Nwith collated and processed imaging data.
This output file mirrors the registration output but with all z-planes collated into a single dataset.
Hint
In the filename, you’ll see min_snr followed by a number. This is also stored in the metadata and is the primary variable dictating the threshold of detection.
In the resulting filename you will see the collated minSNR value. This new file
holds a concatenated, centered and thresholded master copy of all neurons, footprints and traces.