Projection Images#
Suite2p computes several reference images during registration and detection. These images are used for quality assessment and as inputs to Cellpose anatomical segmentation.
Processing Pipeline#
Images stored in ops.npy are computed at different stages:
Step |
Description |
Disable |
|---|---|---|
Temporal Binning |
Average consecutive frames to reduce noise |
|
Temporal High-Pass |
Remove slow baseline drift from movie |
|
* The MBO fork allows disabling high_pass; standard Suite2p always applies it.
What’s stored:
meanImg- mean of binned movie (computed before HP filter)meanImgE- enhanced mean from registration (median filter based)max_proj- max of binned + HP filtered movieVcorr- correlation map or cellpose input image
Anatomical Detection Modes#
Mode |
Image |
Description |
|---|---|---|
|
|
Activity relative to baseline |
|
|
Temporal mean |
|
|
Enhanced mean (recommended) |
|
|
Maximum projection |
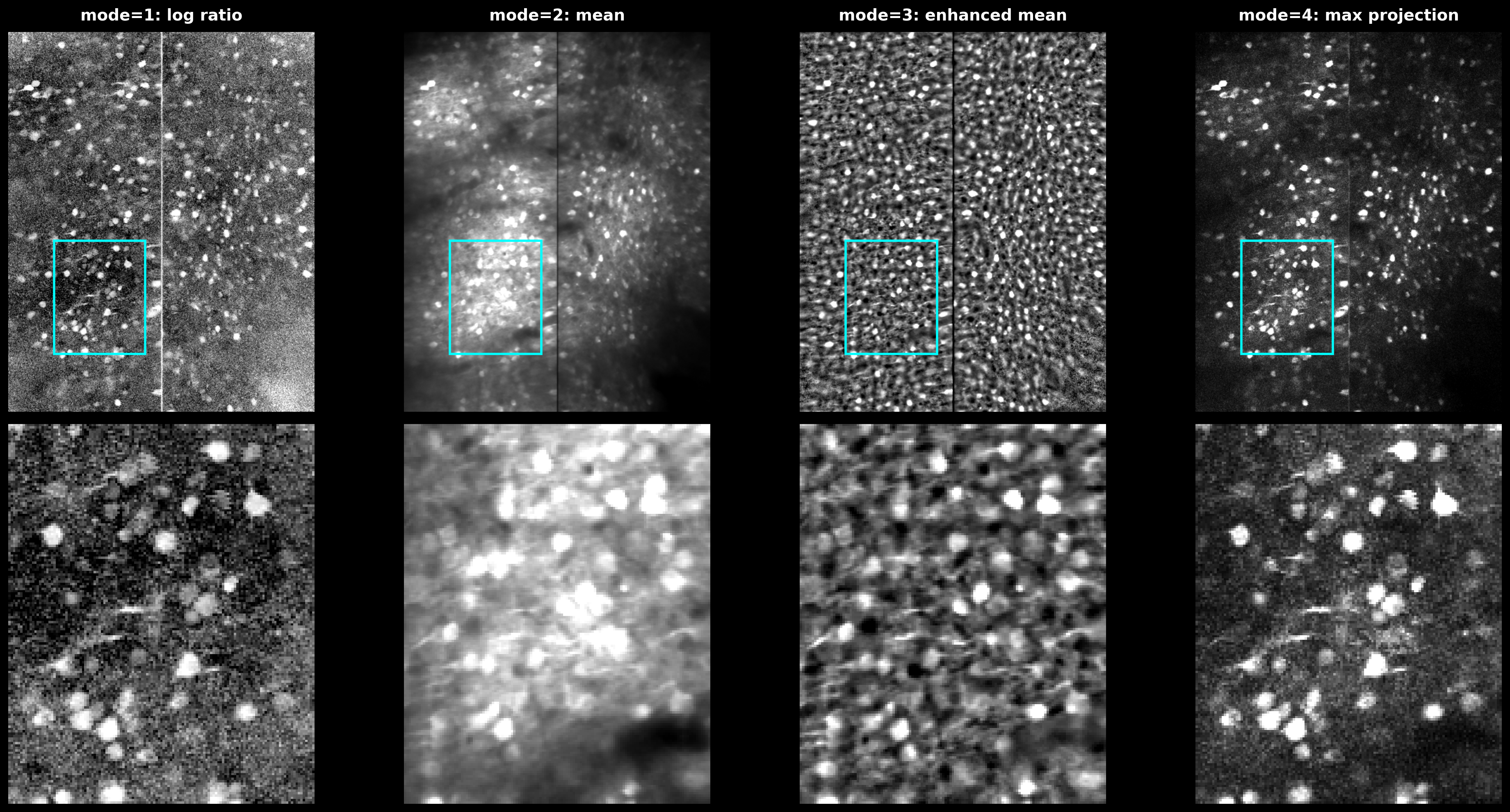
Comparison of anatomical detection modes. mode=1: log ratio highlights active regions. mode=2: temporal mean. mode=3: enhanced mean with median filter. mode=4: maximum projection of HP-filtered movie.#
# mode 1: activity relative to baseline
img = np.log(np.maximum(1e-3, max_proj / np.maximum(1e-3, mean_img)))
# mode 2: temporal mean (before HP filter)
img = mean_img
# mode 3: enhanced mean with local median subtraction
from scipy.ndimage import median_filter
Imed = median_filter(mean_img, size=d)
img = (mean_img - Imed) / median_filter(np.abs(mean_img - Imed), size=d)
# mode 4: max of HP-filtered movie
img = max_proj
Spatial High-Pass Filter#
The spatial_hp_cp parameter applies additional high-pass filtering before Cellpose segmentation. This is separate from the temporal high_pass used during detection.
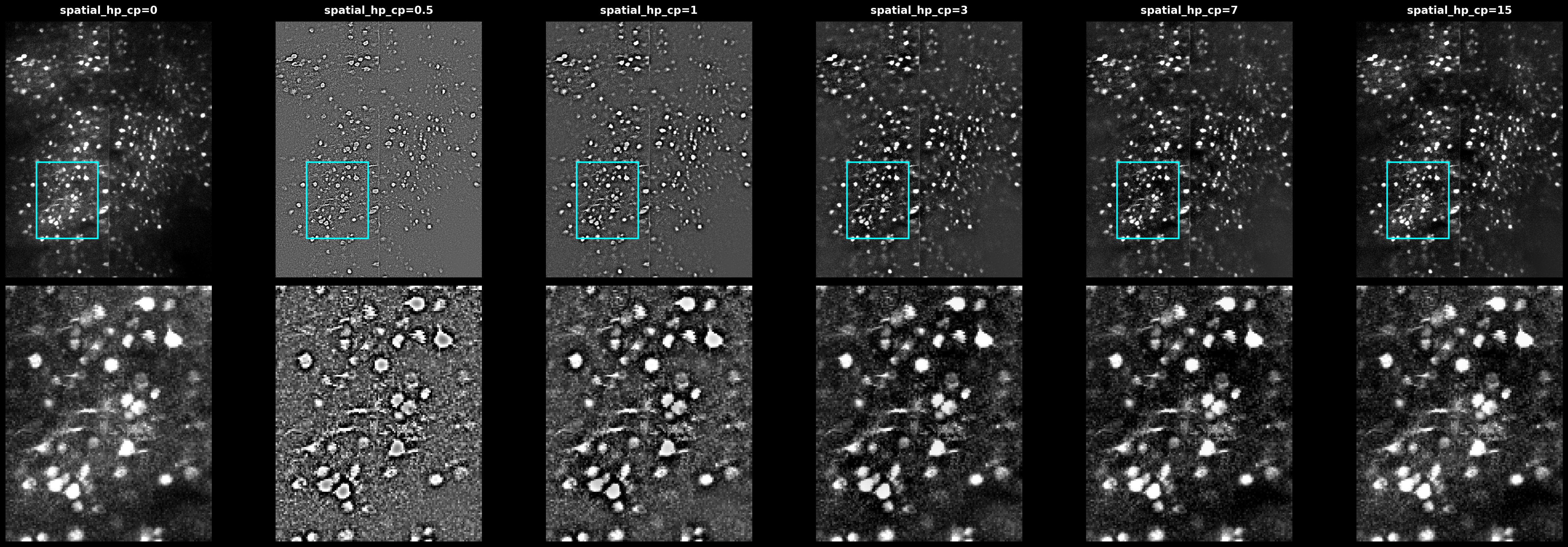
Effect of spatial_hp_cp values on the max projection. Higher values sharpen cell boundaries but may amplify noise.#
from scipy.ndimage import gaussian_filter
def apply_hp_filter(img, diameter, spatial_hp_cp):
sigma = diameter * spatial_hp_cp
return img - gaussian_filter(img, sigma)
Recommendations#
ops = {
"anatomical_only": 3, # use enhanced mean
"spatial_hp_cp": 0, # usually not needed with meanImgE
"diameter": 6, # cell size in pixels
"cellprob_threshold": 0.0,
"flow_threshold": 0.4,
}
If detection is poor:
Try
anatomical_only=4(max projection)Increase
spatial_hp_cpto 1-3Adjust
diameterto match cell sizesTry
anatomical_only=1for data with strong baseline fluorescence
See Also#
User Guide - Complete parameter reference
Cellpose Documentation - Cellpose model details
